Fun with Self-Avoiding Walks Simulations
This post is about some simulations I did of self-avoiding random walks. These are what they sound like: with each step, the position of the walk moves randomly, with the constraint that it can’t visit the same spot more than once. These are mathematically somewhat interesting and crop up in a few areas of physics; I was simulating them because I thought they would solve an unknown problem in polymer physics that I had encountered in my PhD.
Table of Contents
Key Points
- Self–avoiding random walks are simulations that move randomly but don‘t visit the same spot more than once
- The Flory free energy combines entropic elasticity and excluded volume to calculate the excess energy cost of not having particles overlap
- The ideal chain grows as ##R \approx L^{0.5}## and the swollen chain grows as ##R\approx L^{0.6}##
- Semi–flexibility can interfere with excluded volume by making it less likely for two nearby segments of a molecule to bump into each other, and this changes the statistics of the chain
- The blob model says that a chain can be imagined as a 2D self–avoiding walk of spherical blobs, inside each of which the chain does a 3D self–avoiding walk
- As you cram DNA in narrower and narrower slits, the molecule spreads out more and more according to the negative quarter–power of the height of the slit
- My simulation of self–avoiding random walks approaches the asymptotic behaviour of the self–avoiding walk
- Plugging in some of the numbers from my simulations, I can predict a scaling of –0.2 instead of –0.25, closer to the experimental result of 0.16
The Simulation
The simulations work as follows: on a square lattice, starting at the origin, take a step in a random direction. If that direction is occupied, try another direction. If all directions are filled, stop. Aficionados of the video game Snake may recognize this halting condition. The shortest distance it can travel before trapping itself is eight steps (this happens in the gif if you watch carefully), and the average number of steps they travel before stopping is about 70. The main parameters I was measuring were the distance between the two ends, and the radius of gyration, which is essentially the standard deviation of the position of each site (the centre of mass is the mean of each site, the radius of gyration is the root-mean-square displacement of each site from the centre of mass). I was interested in how the radius of gyration grows with the number of steps.
The Wikipedia page gives some examples of their applications in physics and math. An interesting one is that it’s equivalent to the Ising model (interacting magnetic spins on a lattice) with zero degrees of freedom. I have also read a paper comparing the enumeration of self-avoiding walks to the entropy of a black hole surface, which I’d like to learn more about. There’s also an entire book on the subject. I want to mainly discuss them in the context of polymer physics.
A polymer is a long stringy molecule, and the simplest way to model them is as a random walk through space. My second blog post introduces a few of the physical phenomena that polymer chains exhibit, which typically involves maximizing entropy by contracting into blobs. The “ideal chain” model of a polymer has no interactions between different segments of the chain, and no long- or short-range correlation in orientation along the chain, which is why it is described by the random walk. The average distance travelled by a random walk is zero, and the average distance between the two ends of a polymer is zero. However, the mean-square displacement (MSD) of a random walk is non-zero, the mean-square end-to-end distance is non-zero. The MSD is proportional to the number of steps in the walk, so the root-mean-square (RMS) displacement is proportional to the square root of the number of steps. For an ideal polymer, the end to end distance and radius of gyration is proportional to the square root of the length of the chain. In the language of scaling laws, ##R_{g}\approx L^{0.5}##.
 |
| This polymer on a surface totally looks like a random walk! From JACS 124, 13454. |
However, in reality, no two segments of a polymer chain can occupy the same point in space. This means, instead of being described by the random walk, it is described by the self-avoiding random walk. Because the self-avoiding walk excludes configurations that visit the same site, it is generally bigger than a self-avoiding walk of the same length: if you “turn on” self-avoidance interactions, the chain “swells” so this is called a swollen chain. So, if the ideal chain grows as ##L^{0.5}##, how does the swollen chain grow with length?
If we consider some number N particles each with volume v inside a larger volume V, the probability of any two particles occupying the same space is
$$p_{1}=\frac{v}{V}$$
So the probability of them not occupying the same space is:
$$\overline{\,p_{1}}=1-\frac{v}{V}$$
Now, if there are N of these particles, there are N(N-1)/2 pairwise combinations of them, so the probability of no two particles occupying the same space is:
$$\overline{\,p_{N}}=\left(1-\frac{v}{V}\right)^\frac{N(N-1)}{2}$$
Rewriting this and then assuming that N is big, we have
$$\overline{\,p_{N}}=exp\left[\frac{N(N-1)}{2}\log{\left(1-\frac{v}{V}\right)}\right] \approx exp \left[-N^{2}\frac{v}{V}\right]$$
The approximation arises from the Taylor expansion of log(1-x) as well as the fact that N-1 is roughly N. I also dropped a factor of two, because it makes me feel powerful to do that. Now, we take a lesson from the canonical ensemble and remember that according to the Boltzmann distribution, the probability of observing the system in some state is exponentially decaying with the energy of that state. Our probability is already exponentially decaying, so it tells us the excess energy cost of not having the particles overlap due to excluded volume (EV):
$$E_{EV}\approx kT\cdot N^{2}\frac{v}{V}\approx kT\cdot N^{2}\frac{v}{R^3}$$
Here, kT is the thermal energy, and I made an assumption that we live in three dimensions, which isn’t a necessary one. This is half the picture. The other half is entropy: a random walk is more likely to be observed in a high-degeneracy state. A random chain is unlikely to be in a totally extended state; the ends are statistically more likely to be found close to each other. The size of a random walk, being random, has a Gaussian distribution, and through the Boltzmann distribution, this gives rise to entropic elasticity: the energy of extending an ideal chain is harmonic, and the spring constant is its average radius of gyration, which is the product of the step size and the number of steps N.
When we combine these two factors, entropic elasticity and excluded volume, what we’re essentially calculating is the excess energy due to the loss of entropy when you exclude states that don’t overlap. This is known as the Flory free energy.
$$E_{F}=kT\left(\alpha \frac{R^2}{N}+ \beta \frac{N^3}{R^3}\right)$$
I’m interested in how R and N scale with one another, so I’ve dropped all other coefficients and embedded them in Greek prefactors. If we take the derivative of the Flory energy and minimize it with respect to the chain radius of gyration, we find:
$$\frac{dE}{dR}=kT\left(2\alpha \frac{R}{N}-3\beta \frac{N^3}{R^4}\right)=0 \rightarrow R \approx N^{\frac{3}{5}}$$
So, we have derived the scaling relation of the swollen chain. The ideal chain grows as ##R \approx L^{0.5}## and the swollen chain grows as ##R\approx L^{0.6}##. After 10 steps, the swollen chain will be 25% bigger, after 1000 steps it will be twice as big. Through some trickery involving the renormalization group that I don’t understand, the exponent is closer to 0.588 rather than 0.6. You can also replace the three-dimensional volume ##R^3## with some arbitrary dimension ##R^d## to get a more general result. For two dimensions, in which I did my simulations, it is 3/4 instead of 3/5.
Neat derivation, but why did I do those simulations I showed at the beginning? My PhD research was part of a field that uses DNA molecules to test the predictions of polymer physics. An early experiment in this field looked at the diffusion coefficient of different sized DNA molecules and found that the growth exponent was experimentally consistent with 0.588, the wormlike chain. However, there’s an additional effect I haven’t talked about called semi-flexibility, which has to do with the fact that polymers are rigid over short distances. For synthetic polymers, this is typically a length so short as to be negligible, but for DNA it’s about 50 nanometers, much larger than the double-helix diameter of about 2 nanometers. Semi-flexibility can interfere with excluded volume by making it less likely for two nearby segments of a molecule to bump into each other, and this changes the statistics of the chain. A general gap of knowledge in polymer physics is what happens when you combine semi-flexibility with excluded volume.
Is DNA a good model polymer?
In 2013, the ungoogleable Douglas Tree and some co-authors (including my current supervisor) wrote a paper called “Is DNA a good model polymer?” and according to Hinchcliffe’s Rule you can guess what the answer is. Basically, they did simulations of semi-flexible chains with excluded volume; essentially a much more nuanced version of the gif I’ve shown on top, in three dimensions. They measured how the end-to-end distance depends on length for a swollen semiflexible chain and looked at the logarithmic derivative of this trend, which is essentially the “local” Flory exponent at that particular length. For short chains, because they are effectively rigid, they grow linearly in separation with length, and the Flory exponent is close to 1.0 for the shortest chains. For very long chains, they act like self-avoiding walks with a Flory exponent asymptotically approaching 0.588. However, for intermediate length chains, things get tricky, because it’s some combination of short-chain rod behaviour, long-chain excluded volume behaviour, and the fact that medium-length chains are less likely to bump into each other than long ones. In fact, the DNA that people tend to use for experiments, which comes from viral genomes, is right in this intermediate range of lengths where no limiting theory really works.
This was the most interesting paper I read that year, and it really made me think because my research was trying to verify all these theories that assume infinitely long chains. Some aspects of my data didn’t really agree with the theory, so was my data wrong, or was the theory wrong because the molecules it was attempting to describe were too short?
One aspect of this involved DNA confined in a slit smaller than its radius of gyration, narrow enough to make the molecule behave as if it were in two dimensions. There have been several experiments looking at DNA confined in slits and seeing how the “spread” of the molecule depends on the height of the slit.
 |
| DNA spreads out more in narrower slits. Images from here, cartoons by me. |
The theory that predicts the spread of a chain in slit confinement basically works by renormalizing the chain into spheres that fill up the slit, called blobs. Inside each blob, the chain undergoes a self-avoiding walk in three dimensions, with a Flory exponent of 0.6. The renormalized chain of blobs undergoes a self-avoiding walk through the slit in two dimensions, with a Flory exponent of 0.75. You can figure out how big the two-dimensional random walk is by figuring out how much of the chain gets repackaged into each blob, and how many blobs there are. So, if the blob is the same size as the height of slit, h, the number of blobs is the total chain length divided by the length of chain in each blob, and the length of chain in each blob obeys 3D Flory statistics, you can figure out that:
$$R(h)\approx h^{1-\frac{0.75}{0.6}}\approx h^{-0.25}$$
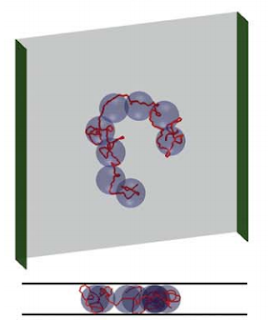 |
| The blob model says that a chain can be imagined as a 2D self-avoiding walk of spherical blobs, inside each of which the chain does a 3D self-avoiding walk. Image from here. |
This is saying that as you cram DNA in narrower and narrower slits, the molecule spreads out more and more according to the negative quarter-power of the height of the slit. The problem was that experiments testing this did not find a result that agreed with this prediction (or with each other, but that’s a different issue).
My idea to resolve this discrepancy was that the chains weren’t long enough to behave like a 2D self-avoiding walk, as Tree and friends showed in 3D. There is a good reason to suspect this because if you look at pictures of DNA in narrow slits, you will see that it doesn’t really encounter itself that often. So, inspired by the 3D simulations of semi-flexible self-avoiding walks, I wrote my simulation of 2D self-avoiding walks.
 |
| Let’s have another look. |
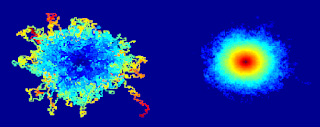
If you look at the aggregate of 10,000 iterations of this algorithm, each of which runs until the molecule traps itself, it looks like the Flying Spaghetti Monster. If you look at a probability density plot of how likely each site is to be visited, it looks like a volcano or the Eye of Sauron.
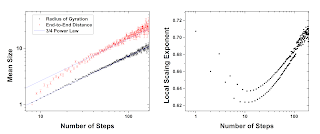
For each step of the self-avoiding walk, I calculated the radius of gyration and the end-to-end distance and kept track of the data as a function of the number of steps. Looking at how these parameters grow with length, we generally recover the 3/4 scaling predicted by Flory theory (a power-law fit gets 0.72).
However, if we look at the logarithmic derivative, d log R/d log N, we can see how it approaches this asymptotic limit with length. It is similar to the 3D case. Starting from the rod-like limit (when you go from one to two steps, it can only double in size) it falls towards the ideal 0.5, reaching a minimum and then rising towards 0.75. My data isn’t quite enough to reach that limit, and there’s this weird even-odd pattern which I believe is caused by the fact that I discretize the walks to a square lattice. Because walks longer than length 70 get relatively rare, the noise in the simulation picks up after that point.
This possibly suggests a correction to the standard Flory prediction for a molecule’s size in a slit, but the problem is that the same derivation doesn’t really work because it assumes that these scaling relations hold, and you can’t really change some of the numbers without really changing the whole form of the equation. Plugging in some of the numbers from my simulations, I can take the minimum exponent which is about 0.65 and the minimum 3D exponent from Tree’s paper, which is about 0.54, I predict a scaling of about -0.2 instead of -0.25. This is closer to the experimental result of about 0.16 but still doesn’t fit the data, and isn’t really physically justified.
Conclusion
I stopped working on this particular research avenue shortly after figuring this out, but it was kind of fun creating this random walk simulation and seeing how it approaches the asymptotic behaviour of the self-avoiding walk. This was a fairly long and rambly story about why I was doing these simulations. Just to summarize, I started simulating self-avoiding random walks because I thought that an unknown aspect of their behaviour, the asymptotic growth of their Flory exponent, would resolve some standing questions in polymer physics. It didn’t, but I tried.
Ph.D. McGill University, 2015
My research is at the interface of biological physics and soft condensed matter. I am interested in using tools provided from biology to answer questions about the physics of soft materials. In the past I have investigated how DNA partitions itself into small spaces and how knots in DNA molecules move and untie. Moving forward, I will be investigating the physics of non-covalent chemical bonds using “DNA chainmail” and exploring non-equilibrium thermodynamics and fluid mechanics using protein gels.








With a back-tracking algorithm, it should be possible to list all cases (less than 30,000) and their probabilities. The less than 1 million cases for 14 steps should still work, afterwards it might need some computing time. n steps in a self-trapping walk need at most n-5 empty space in every direction from the starting point, but grid size should not be a limit anyway.
If we define the first step to be North, we have 10 steps left for the 11-step walk, removing the mirror symmetry each path has a probability of at least 2/3^10 or 1 in 30,000. With an expected occurrence of 30 or more in a million samples, it is extremely unlikely that you missed any pattern. It should be possible to automatically analyze those patterns in terms of the number of available spaces in each move, which leads to the probability of this path.
p { margin-bottom: 0.1in; line-height: 120%; }mfb -Thank you for explaining how to count the probabilities for each path of a random walk! And for soothing my pride with the word “tricky”. I have found six 9-step trapped walk paths that end next to the starting square. Together with their mirror images that makes twelve. I also found four 9-step paths that end a knight’s move from the starting square and one that ends three steps from the starting square in the direction of the first step. With their mirror images that brings the total number of 9-step paths to 22. I have worked out the probabilities for the 9-step walks and they check nicely with my simulation.That led me to try the 10-step case where I found 25 paths with their mirror images to make 50 total. Again, the calculated and simulated results agree; see attached graph. I found at least 190 different 11-step trapped walk paths (including symmetric ones) in a sample of 10^6 walks. The samples of 11-step paths ranged in size from 93 down to 10.I have a copy of “The Self-Avoiding Walk” by Madras and Slade on order in hopes it may provide further insight into this system.
I found at least 190 different 11-step trapped walk paths (including symmetric ones) in a sample of 10^6 walks. The samples of 11-step paths ranged in size from 93 down to 10.I have a copy of “The Self-Avoiding Walk” by Madras and Slade on order in hopes it may provide further insight into this system.
[QUOTE="herb helbig, post: 5564985, member: 602887"]I suppose, more accurately, the 8-step probability should be reduced by multiplying by the probability of not being trapped at 7 steps: 1-P(7) = 0.99726, but it would take a simulation of about 50 million walks to verify that, and much confidence in the random number generator![/QUOTE]The probabilities are absolute. If we would have those values for all N, they would add to 1.The 8-step path has a tricky case: if you go NNWWSSE (using the directions as you would expect), there are just two instead of three options left for step number 8, one of them ends.Define North to be the direction of the first step.- With 1/3 probability the next step is also N, then we have 2/3 probability to go E or W, afterwards we have to choose the right direction out of 3 four times in a row, and finally a 1/2 chance to get trapped. Total: 1/3[sup]6[/sup] = 3/3[sup]7[/sup]- With a 2/3 probability the next step is W or E, then the next 6 steps have to pick the right direction out of 3, for a total contribution of 2/3[sup]7[/sup].Sum: 5/3[sup]7[/sup]The 7-step walk has 1 case, the 8-step walk has 2 cases, the 9-step walk has 6 cases, and the 10-step walk has at least 15 cases. Manual analysis becomes messy quickly.[QUOTE="herb helbig, post: 5564985, member: 602887"]The trend of your (n+1)/2, N/2 parity effect is evident in the data, but apparently an underestimate.I have a figure summarizing what I have said, but not sure whether it has uploaded.[/QUOTE]That (n+1)/2 vs n/2 pattern should have an odd/even pattern.
[QUOTE="mfb, post: 5559051, member: 405866"]I see an effect from parity (like black/white checkerboard fields), but I would not expect it to be so large: a walk after an odd number of steps N can potentially be blocked by (N+1)/2 other elements, while a walk after an even number of steps can be blocked by N/2 other elements.The probability of a walk ending after 7 steps is 6/3[sup]7[/sup], while the probability of ending after 8 steps is 5/3[sup]7[/sup].[/QUOTE]mfb – Again many thanks for taking an interest in this problem. The wheels in my 82 year old head don't spin as fast as they once did! Your values for the probabilities, P(7) and P(8), of 7 and 8 step walks agree well with two 1-million walk simulation runs;P(7)=6/3^7 Run 1 Run 20.002743 0.002755 0.002791P(8)=5/3^70.002286 0.002237 0.002258I think I understand 6/3^7; each step has 3 branches so in 7 steps there are 3^7 possible walks, and for each of the 3 branches there is a left- and a right-handed trapped loop making 6 successes. (Since trapping involves a curling path, is it so that all trapped walks come in left- and right-hand pairs?)Attempting to use the previous reasoning for the 8 step walk leads me to this (wrong) conclusion: 3 branches for 8 steps gives 3^8 walks, for each branch there is the same, left/right pair of 7-step trapped loops, but with two possible additional first steps for a total of 12 successful 8-step walks. That gives me 12/3^8 = 4/3^7 instead of your 5/3^7. I have spent many waking and even dreaming hours trying to understand why 4 must be 5! I'm sure I'm missing something that should be obvious, but would be grateful for your explanation!I suppose, more accurately, the 8-step probability should be reduced by multiplying by the probability of not being trapped at 7 steps: 1-P(7) = 0.99726, but it would take a simulation of about 50 million walks to verify that, and much confidence in the random number generator!I start all walks with the same [1,0] step since symmetry suggests that this should have no effect on the distribution of walk lengths, and it will allow me to see what effect this bias might have on final displacement.The trend of your (n+1)/2, N/2 parity effect is evident in the data, but apparently an underestimate.I have a figure summarizing what I have said, but not sure whether it has uploaded.
I see an effect from parity (like black/white checkerboard fields), but I would not expect it to be so large: a walk after an odd number of steps N can potentially be blocked by (N+1)/2 other elements, while a walk after an even number of steps can be blocked by N/2 other elements.The probability of a walk ending after 7 steps is 6/3[sup]7[/sup], while the probability of ending after 8 steps is 5/3[sup]7[/sup].
[QUOTE="mfb, post: 5558877, member: 405866"]A walk ends if there is no free spot any more?Up to symmetries, there is just one 7-step walk (up, left, left, down, down, right, up), and two 8-step walks (same pattern starting one step later). The 8-step walk needs a more precise arrangement of the steps.[/QUOTE]Yes, thank you for the prompt reply! But isn't it odd that this bias should persist for all walks up to a length of 50 steps or more? And if that sort of argument is valid, how does it apply to, for example, channels 8 and 9? Or, for that matter, channels 48 and 49 where the distribution is past its maximum (which occurs at ~ walks of length 30 steps?
A walk ends if there is no free spot any more?Up to symmetries, there is just one 7-step walk (up, left, left, down, down, right, up), and two 8-step walks (same pattern starting one step later). The 8-step walk needs a more precise arrangement of the steps.
I've written julia code (happy to share) to simulate self avoiding random walks on a square grid, and collected data on the distribution of walk lengths (number of walks of length L). The distribution shows an average walk length of 70.66 steps in good agreement with Hemmer and Hemmer (J. Chem. Phys. 81, 584 (1984); http://scitation.aip.org/content/aip/journal/jcp/81/1/10.1063/1.447349). What surprised me was that adjacent columns of the frequency histogram were staggered in height, the odd columns being larger than the even from the shortest walk (7) at least up to walks of length 50 steps. Over this range, the odd walk numbers are at least 3% higher than the next larger walk number, with channel counts from ~2500 to ~11,000 (so the statistics are sound). For example, the 7-step walk channel had 2755 counts while the 8-step walk channel had 2237 counts. Has anyone noticed this or have an explanation?
[QUOTE=”klotza, post: 5440026, member: 569939″]Hi Jimster41 I just saw your questions now, several months after you asked them.
The self-avoiding walk only grows from the end, it has linear topology in its basic form. Other versions are bottle-brush polymers or comb polymers, hyperbranched polymers, dendritics, etc, which have branchings.
I don’t know much about self organizing criticality and scale-invariance, but I do know that regular brownian diffusion can be described by a fractal wiener process. The mathematical literature on the critical behaviour of SAWs is quite extensive but a bit beyond me.[/QUOTE]
Thank you sir. I know approximately 100% more about pink, white and brown noise, Weirstrauss, Sierpinski, SARW, fractals, self similarity, Lorenz sytems, and the zoo of such things now, having slogged through Manfred Schroeder’s book on them (which by the way reads like a walk across the islands of Galapagos).
Your article was one of the things that prompted me to actually try hard to read that… Unfortunately, as you know, 100% of almost nothing is still… next to nothing.
Not that I’m complaining. It’s great to feel completely baffled.
Hi Jimster41 I just saw your questions now, several months after you asked them.
The self-avoiding walk only grows from the end, it has linear topology in its basic form. Other versions are bottle-brush polymers or comb polymers, hyperbranched polymers, dendritics, etc, which have branchings.
I don’t know much about self organizing criticality and scale-invariance, but I do know that regular brownian diffusion can be described by a fractal wiener process. The mathematical literature on the critical behaviour of SAWs is quite extensive but a bit beyond me.
In the growth of a real polymer can the “walk” take its next step from more than one location on the existing molecule or is it always a literal chain?
Is there evidence of Self Organizing Criticality and Scale-Invariance in a SARW or in real polymers? The picture of Sauron made me wonder.
I am struck ([I]and confused by[/I]) the similarity between a SARW and an Abelian Sandpile with a few rule changes – but then I guess there are lots of variations on such games.
Nice article.
The probability estimate for multiple elements at the same point seems to assume that the positions are completely uncorrelated. That is certainly not true for a random walk, you have strong correlations between neighbor steps. Is there some justification that those correlations don’t change the result.
[QUOTE=”klotza, post: 5262780, member: 569939″]klotza submitted a new PF Insights post
[URL=’https://www.physicsforums.com/insights/fun-self-avoiding-walks/’]Fun with Self-Avoiding Walks[/URL]
[IMG]https://www.physicsforums.com/insights/wp-content/uploads/2015/10/animationwalk-80×80.png[/IMG]
[URL=’https://www.physicsforums.com/insights/fun-self-avoiding-walks/’]Continue reading the Original PF Insights Post.[/URL][/QUOTE]
“The size of a random walk, being random, has a Gaussian distribution” I would think it would be a Poisson distribution. In this circumstance it would approximate a Gaussian though.
Hi Jimster41 I just saw your questions now, several months after you asked them.The self-avoiding walk only grows from the end, it has linear topology in its basic form. Other versions are bottle-brush polymers or comb polymers, hyperbranched polymers, dendritics, etc, which have branchings.I don't know much about self organizing criticality and scale-invariance, but I do know that regular brownian diffusion can be described by a fractal wiener process. The mathematical literature on the critical behaviour of SAWs is quite extensive but a bit beyond me.
Great article! Useful animations!