When did Mitochondria Evolve?
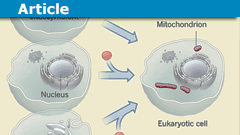
Historically, life is categorized into three broad domains: bacteria, archaea, and eukaryota. Whereas bacteria and archaea are almost all relatively simple, single-celled microorganisms, humans and all other forms of complex, multi-cellular life (including all other animals, plants, and fungi) are classified as eukaryotes, so understanding the evolution of eukaryotes is important for understanding the origin of cellular complexity. Eukaryotes have a number of unique features—including a nucleus (which separates the genetic material from the rest of the cell), an endomembrane system (a system of lipid membranes inside the cell which facilitates the transport of materials and enables the cell to compartmentalize many functions), and mitochondria (which aid in energy production and metabolism)—that are thought to underlie their greater complexity. However, how all of these features evolved is still unclear.
Recent research has shown that rather than being a separate domain of life, eukaryotes actually represent the fusion of an archaeal host and a bacterium that eventually became the mitochondria, through a process called endosymbiosis. This event led to the integration of many bacterial genes into the genome of eukaryotes such that the eukaryotic genome contains a mixture of informational genes from archaea (involved in processes such as DNA replication, RNA production, and protein synthesis) and operational genes from bacteria (involved in metabolism). Thus, the evolution of mitochondria was a key step in the evolution of eukaryotes.
However, understanding the role of mitochondria in the evolution of cellular complexity requires understanding when in the process of eukaryotic evolution mitochondria were acquired. Researchers have speculated that the evolution of mitochondria was the first step in the evolution of eukaryotes (the mito-early hypothesis), and that the acquisition of mitochondria provided the energy production capacity to drive cells to encode more genes, synthesize more proteins, and make bigger cells overall. Indeed, because all known eukaryotes either have mitochondria or show evidence that they had mitochondria in the past, researchers have generally favored the mito-early hypothesis. However, a recent paper published in Nature argues against the mito-early hypothesis and finds genetic evidence that mitochondria were acquired later than the genes involved in the endomembrane system.
The authors of this study analyzed the genomes of many eukaryotes in order to get a sense of what the genome of the last common ancestor of all eukarotes looked like. Then, for each gene in the eukaryotic common ancestor, they compared that gene to related genes from archaea and bacteria in order to determine whether the gene originally came from an archaeon or bacterium. Finally, they determined the number of mutations accumulated between the eukaryotic genes and their archaeal or bacterial ancestors to estimate how long ago eukaryotes acquired these genes.
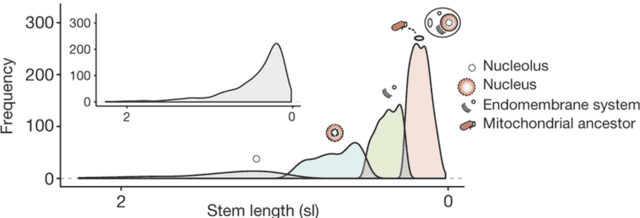
As expected, the most ancient sets of genes in the eukaryotic genome are all archaeal in origin, reflecting the fact that the host cell in endosymbiosis was an archaeal cell. The genome also contains a set of genes related to alphaproteobacteria, which are the genes introduced into the genome during the acquisition of mitochondria. Unexpectedly, the authors identified the third set of bacterial genes in eukaryotes that do not seem to originate from alphaproteobacteria and appear to have been acquired prior to the mitochondrial genes. Many of these genes seem to function in the endomembrane system, suggesting that this feature predated the evolution of mitochondria. The distinction is even more striking when the authors used computational methods to group the genes by evolutionary age (referred to as the “stem length” in the figure. Shorter stem lengths, which are further to the right on the plot, indicate more recent acquisition of these genes):
This computational grouping method identified four groups, and the four groups seem to be associated with different cellular functions. The two most ancient groups of genes, involved in the nucleus and a substructure of the nucleus called the nucleolus, are primarily of archaeal origin. The next most ancient is the aforementioned group associated with the endomembrane system, which appears to be older than the youngest group of genes that originated from the mitochondrial ancestor. Overall, the genetic data suggest that the evolution of the endomembrane system predated the acquisition of mitochondria.
Of course, as the argument is based primarily on genetic data, it would be wise to wait for these findings to be confirmed by other techniques before declaring the question solved. Indeed, the findings have generated some controversy among evolutionary biologists as analyzing DNA sequence data requires making a number of assumptions. In particular, it can be tricky to estimate age from the accumulation of mutations as the authors of this paper has done. The number of mutations a gene accumulates not only depends on the time since the gene diverged from its ancestor but also on the rate of mutation. If the genes involved in the endomembrane system had to undergo many more changes so to adapt to the environment of the eukaryotic cell than the mitochondrial genes (a plausible hypothesis is given the differences between bacterial and eukaryotic cell membranes), it could still be possible that the genes involved in the endomembrane system were acquired later than or around the same time as mitochondria.
Instead, finding an evolutionary “missing link”—an eukaryotic-like organism containing an endomembrane system but lacking mitochondria—would provide slam-dunk evidence for the mito-late or mito-intermediate hypotheses. Work on the newly discovered Lokiarchaeota phylum of archaea could potentially provide this evidence as these organisms possess some genes that hint at the presence of an endomembrane system.
If correct, disproving the mito-early hypothesis would change our prevailing theories about the origin of eukaryotes and the role of mitochondria in their evolution. Even if the mito-early hypothesis ends up being correct, however, the study still raises many interesting questions about the origin of the endomembrane system and suggests that multiple waves of bacterial genes made their way into the eukaryotic genome. Thus, despite the remaining uncertainty, the study by Pittis and Gabaldón provides a good argument to reconsider the mito-early hypothesis and advances our understanding of the evolution of cellular complexity.
Further Reading:
Pittis and Gabaldón. 2016. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature. Published online 3 February 2016. doi:10.1038/nature16941
Ettema 2016. Evolution: Mitochondria in the second act. Nature. Published online 3 February 2016. doi:10.1038/nature16876
Postdoctoral researcher in biophysics.
My areas of expertise include single molecule spectroscopy, structural biology, biochemistry, and evolutionary biology.





UPDATE: Please see the response to the critique.
[URL]http://treevolution.blogspot.ca/2016/05/response-to-late-mitochondrial-origin.html[/URL]
UPDATE: William Martin and co-authors have published a (non-peer reviewed) critique of the Pittis and Gabaldón study. They take issue with the use of stem-length (sl) as a measure of evolutionary age and with some of the methods used to analyze the data in the original publication:
[quote]In summary, sl-based conclusions about eukaryote evolution are unfounded, resting upon fatal
flaws in i) over-fitting of the wrong distribution model, ii) analyses of non-independent data,
and iii) implicit, untested, and untrue molecular clock assumptions.[/quote]
Martin et al. 2016. Late mitochondrial origin is pure artefact. bioRxiv doi:10.1101/055368
The full paper is freely available at [URL]http://biorxiv.org/content/early/2016/05/25/055368[/URL]
[QUOTE=”jim mcnamara, post: 5371259, member: 35824″]I’m not current in this field. A priori, an understanding of chloroplast development should help verify the endomembrane when question. Do you have any links on this? Chloroplasts have membranes within membranes – e.g., thylakoid membrane. Cyanobacteria are candidates for a possible endosymbiotic source for chloroplast development.[/QUOTE]
I haven’t looked too in depth into chloroplast evolution or thykaloid evolution in cyanobacteria, but those are good thoughts. Chloroplasts are believed to have evolved after mitochondria through endosymbiosis with a fully eukaryotic host containing nucleus, endomembrane system and mitochondria (here’s a nice [url=http://www.sciencedirect.com/science/article/pii/S0960982208014851]review article[/url] on the evolution of chloroplasts). However, perhaps the beginnings of an endomembrane system evolved in an organism like cyanobacteria and got transfered to the eukaryotic ancestor at some point.
[QUOTE=”Torbjorn_L, post: 5371501, member: 488061″]- I am not surprised after the Lokiarchaeota phylum result. Moreover the usual comparison between prokaryote and eukaryote energy efficiency (such as Lane’s) is problematic. Comparing apples with apples prokaryotes can sustain about as large protein turnover (so large genomes) as eukaryotes. [ [URL]http://book.bionumbers.org/what-is-the-power-consumption-of-a-cell/[/URL] ] And I think there is a paper that directly comes to the same conclusion. [A lost reference as I write this in haste. :-/] So mito-late would presumably be viable.[/quote]
Yes, the hypothesis argued by the Lane paper is controversial. Here’s one criticism of the hypothesis published recently in the [i]Proceedings of the National Academy of Sciences[/i]: [URL]http://www.pnas.org/content/112/33/10278.abstract[/URL]
Interestingly, in the supplementary materials of the Pittis and Gabaldón paper, they claim that the group of bacterial-origin genes they identified to be putatively involved in the endomembrane system are not present in the Lokiarchaeota sample.
[quote]- The “controversy” reference is peculiar in criticizing the use of trees by default since coarse history is well captured by them, including the endosymbiosis in question![/QUOTE]
The tree criticism is somewhat valid given that the paper is trying to understand horizontal gene transfer, something that tree models are not designed to handle. It’s possible that imposing a tree model on a more complicated evolutionary process could cause some of the molecular clock estimates to be wrong.
Very useful synopsis, as the paper is on my to read pile!
Some hasty reflections:
– I am not surprised after the Lokiarchaeota phylum result. Moreover the usual comparison between prokaryote and eukaryote energy efficiency (such as Lane’s) is problematic. Comparing apples with apples prokaryotes can sustain about as large protein turnover (so large genomes) as eukaryotes. [ [URL]http://book.bionumbers.org/what-is-the-power-consumption-of-a-cell/[/URL] ] And I think there is a paper that directly comes to the same conclusion. [A lost reference as I write this in haste. :-/] So mito-late would presumably be viable.
– The ER and nucleus has the wrong topology to be inherited vertically as a functional unit. Rather the Lokiarchaeota paper solves this.
– Both the Lokiarchaeota paper and the mito-late result would be consistent with the latest mitochondrion phylogeny (that I know of). Having the mitochondrion ancestor as an energy parasite could mean many infestations before the parasite was captured and defanged by increasing mutualism. [ [URL]http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0110685[/URL] ]
– The “controversy” reference is peculiar in criticizing the use of trees by default since coarse history is well captured by them, including the endosymbiosis in question!
Very well done write up. It triggered some some thoughts related to chloroplast evolution. Thanks for a nice job.
I’m not current in this field. A priori, an understanding of chloroplast development should help verify the endomembrane when question. Do you have any links on this? Chloroplasts have membranes within membranes – e.g., thylakoid membrane. Cyanobacteria are candidates for a possible endosymbiotic source for chloroplast development.
Great as always!